Microsatellite repeat expansion disorder is a class of disorder which was discovered in 1991, in the fragile X mental retardation 1 (FMR1) gene, caused due to the expansion of a CGG repeats. Until now, nearly 40 such microsatellite repeat expansion disorders have been identified. Such an expansion results in an unusual DNA and RNA conformations and leads to several neurological and neurodegenerative disorders. There is no proper cure available for these disorders until now. To this end, our lab focuses on the unusual DNA and RNA conformations taken by such microsatellite repeat expansion disorders and their consequences in interaction with proteins. Our lab also focuses on identifying lead molecules to treat such disorders by considering DNA and RNA as the targets. We employ sub-cloning, expression, EMSA, CD, NMR and molecular dynamics simulations to address this challenging problem.
Collaborators: Prof. Prabu Shankar (Department of Chemistry, IITH), Dr. Subha Narayan Rath ( Department of Biomedical Engineering, IITH) and Dr. Krishna Rao (TCIS, Hyderabad).
Multidrug resistance in Gram-negative bacteria is an important challenge the world is witnessing now. Frequent outbreak of such multidrug resistance in Gram-negative bacteria has become a health threat. In this context, using E. coli, Kebsiella spp., Acinetobacter spp. etc. as model systems we try to utilize the capsular polysaccharide (CPS or K-antigen) surface exportation nanomachinery as a drug target to treat these bacterial infections Techniques employed are sub-cloning, expression, EMSA, CD, NMR and docking
Extension of this project is the in-silico serotyping of these Gram-negative bacterial species using the proteins involved CPS surface exportation to facilitate the outbreak surveillance and disease epidemiology Yet another extension, is modeling and creation of three-dimensional database of CPS or K-antigen of these bacterial species.
Following the SARS-CoV-2 pandemic outbreak in 2019, our laboratory explored the evolutionary tactics followed by SARS-CoV-2 through the pan proteomic/genomic analyses. We have also addressed
(i) zoonotic and zoo-anthroponotic transmissions and the concomitant evolution of the virus during the pandemic
(ii) the role of using SARS-CoV-2 environment sequence as the tool to monitor the viral distribution in the community
(iii) the need of epitope-based vaccine design to tackle the virus
Importantly, we have launched an interactive database to monitor SARS-CoV-2 evolution. Methods employed in this project are: sequence analysis, phyloproteogram, immunoinformatics and in-house bash/python/html/mysql scripting.
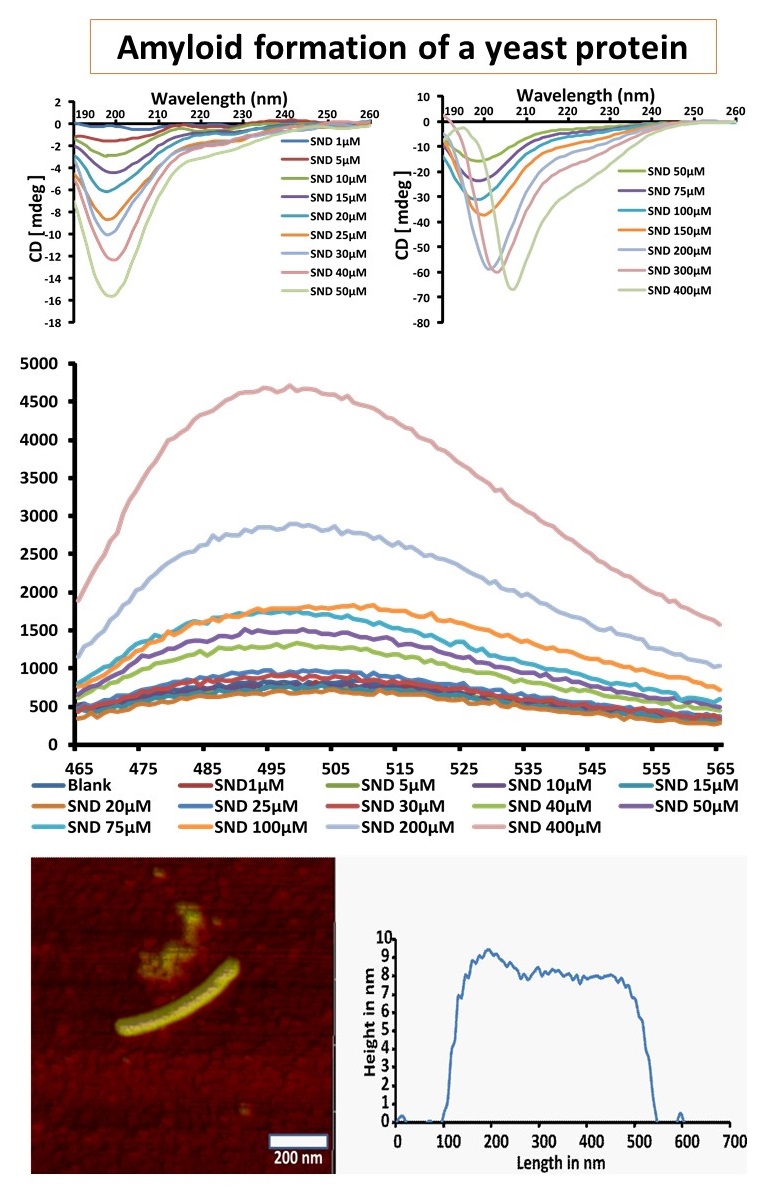
Demonstration of the ability of a Saccharomyces cerevisiae protein to form amyloid by employing sub-cloning, protein expression, CD, Th-T and AFM techniques. Further, understanding the mechanism of amyloid formation using mutagenesis.
Our lab also extensively involved in developing application tools which can be useful for scientific community. This involves, 3D-NuS (automated nucleic acids modelling server), CD-NuSS (a server uses machine learning approach to predict the nucleic acids secondary structures using CD spectra), STRIDER (an application tool to estimate the steric hindrance of a biomolecule/complex), K-PAM (Klebsiella spp. serotype predictor), ABSD (Acinetobacter serotyper and surface antigen database), EK3D (Escherichia coli K-antigen three dimensional database), ZnF-Prot (a server for proteome wide prediction of ZnF motifs), CoVe-tracker (an interactive database of SARS-CoV-2 proteome/genome evolution) and a few more on the way ...
SERBP1 is a RNA binding protein and plays an indirect role in epigenetic regulation. It is found to be upregulated in several cancers, including prostate. Here, Sub-cloning, expression, NMR and computational techniques will be employed to utilize it as a drug target.
Our lab has several collaborative projects regarding in silico and in vitro investigations of nucleic acid structural characterization with Dr Umashankar (IITG) and Dr Umakanta (CSIR-IMMT). We also collaborate with Dr David Fernando Estra, Jacobs School of Medicine and Biomedical Sciences, University of Buffalo on protein MD simulations